Free Online Productivity Tools
i2Speak
i2Symbol
i2OCR
iTex2Img
iWeb2Print
iWeb2Shot
i2Type
iPdf2Split
iPdf2Merge
i2Bopomofo
i2Arabic
i2Style
i2Image
i2PDF
iLatex2Rtf
Sci2ools
106
click to vote
ISBI
2008
IEEE
2008
IEEE
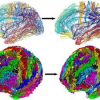
Multi-scale diffeomorphic cortical registration under manifold sulcal constraints
Neuroimaging at the group level requires spatial normalization across individuals. This issue has been receiving considerable attention from multiple research groups. Here we suggest a surface-based geometric approach that consists in matching a set of cortical surfaces through their sulcal imprints. We provide the proof-of-concept of this approach by showing 1) how sulci may be automatically identified and simplified from T1-weighted MRI data series, and 2) how this sulcal information may be considered as landmarks for recent measure-based diffeomorphic deformation approaches. In our framework, the resulting 3D transforms are naturally applied to the entire cortical surface and MRI volumes.
| Added | 20 Nov 2009 |
| Updated | 20 Nov 2009 |
| Type | Conference |
| Year | 2008 |
| Where | ISBI |
| Authors | Alain Trouvé, Arnaud Cachia, Eric Bardinet, Guillaume Auzias, Jean-Francois Mangin, Joan Glaunes, Olivier Colliot, Pascal Cathier, Sylvain Baillet |
Comments (0)